Reports: ND255615-ND2: Roles of Microbial Hydrolytic Enzymes in Mediating Carbon Cycling in Shale Formations of the Southwestern United States
Farooq Azam, PhD, University of California, San Diego (UCSD)
I] Impacts of this grant on:
Research directions in the laboratory of the Principal Investigator and beyond:
My laboratory has been pursuing characterization of microbe-mediated carbon cycling in the marine water column, including micro-scale interactions of diverse microorganisms with a vast repository of dissolved organic matter. While my group has studied proxies of microbial activity in marine sediments, this had not been my focus.
This new directions (ND) grant has enabled me to initiate investigating microbial associations with relatively old and presumably recalcitrant organic matter (OM) of shale formations in the southwestern US. This project is an opportunity to adapt methods as well as understanding gained from ongoing work in my laboratory to the current investigation concerning how microorganisms cycle 'recalcitrant' organic matter within shale bearing sedimentary formations of varying ages. The opportunity to discover the identity of microbial communities mediating C-cycling in such sedimentary formations and understanding some of the underlying molecular-level mechanisms is a strong motivator. This ND-project could help me in formulating new ideas for a subsequent proposal.
This ND project could have relevance beyond the southwestern US and to geological ages of terrestrial shale samples under investigation and broadly they could deepen our understanding of the use of 'recalcitrant' OM by microorganisms. This project could also inform us of microbial potential to hydrolyze analogous shale bearing and other high OM strata in the marine subsurface. These indicate possibilities of learning about potential strategies for utilizing shale bearing sedimentary subsurface microorganisms in releasing tightly bound kerogenic OM from these matrices.
Training and career development:
In keeping with emphasis of this grant on providing a platform for personnel training, I have recruited Dr. Gowtham Subbarao on this project. Dr. Subbarao trained as a protein biochemist during his graduate work focusing on enzyme structure-function relationships and also has experience with biophysical characterization of bacterial membrane proteins. His interest in oceanography motivated him to develop projects on the marine subsurface biosphere. Dr. Subbarao could contribute to this project while also benefiting by:
(i) Acquiring experience with several environmental microbiology procedures including advanced microscopy in my laboratory.
(ii) Developing collaborations with domain experts to further project objectives.
(iii) Participating in fieldwork with experts on shale formations under consideration. This is important for anyone pursuing geo-microbiological work.
This training and experience could serve Dr. Subbarao well as he develops a program on the deep biosphere, especially in the context of emerging energy resources. This project is an excellent vehicle to achieve these objectives. An undergraduate student also interned over this summer on this project, providing a flavor of this multidisciplinary research to him prior to embarking on graduate school.
II] Work progress:
1] We are employing a multi-pronged approach to sample selection and collection from representative field sites spanning geological time. Prof Richard Norris, our collaborator at Scripps, provided a few shale samples (Colorado and Wyoming) from his collection. We have also obtained samples from southern California (Monterrey formation) and high OM bearing sediment cores from the anoxic Santa Barbara basin. We shortly intend collecting additional samples from these sites.
Figure 1: Shale sample from Green river formation, Colorado (GRCO), USA. The shape of this sample approximates a cuboid with approximate dimensions: 7 X 3.5 X 5 inches3.
2] Characterization of microbial diversity in select shale samples:
We are employing complementary approaches of 16S rDNA sequencing & Intact Polar Lipids (IPLs) characterization to unravel microbial diversity of shale sedimentary rocks.
(i) Determining archaeal and bacterial diversity: We extracted genomic DNA from several shale samples to prepare archaeal & bacterial libraries for 16S rDNA sequencing.
(ii) Dr. Subbarao initiated collaborative studies on IPLs with Dr. Julius Lipp at MARUM, Germany to investigate proportion of archaea and bacteria in the studied biosphere associated with shale samples. IPLs also enable determining ratio of fossilized to living microorganisms in samples spanning slices of geological time. These studies are ongoing.
3] Presence and nature of microorganisms in shale matrices: (Dr. Subbarao collaborates on this theme with Nirav Patel, one of my graduate students)
3a] Fluorescence imaging: We employed traditional fluorescence methods to attempt visualizing microorganisms in shale matrices. The topography of surfaces within a shale rock is typically uneven. This makes such samples challenging for epifluorescence microscopy. This has necessitated employing laser confocal microscopy in our laboratory with a variety of dyes that stain different bio-molecular classes on shale chip surfaces (some images are presented below). We visualized fluorescently stained objects that appear to be microbial cells, some of them viable, on select surfaces in the interior of shale rock. Select images are presented here.
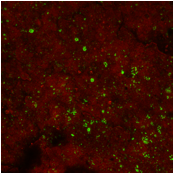
Figure 2 (please see image to the left): SYBR-Green-I stained shale chip surface from GRCO. SYBR-Green-I binds microbial cell sized (< 2.0 µm) structures (DNA) arranged individually or in clusters (see green colored structures). FM-4-64fx with an affinity for hydrophobic regions appears to coat the chip surface in a non-specific manner. Scale used in images in this report: 215 micrometer X 215 micrometer. We are in the process of confirming identities of these green colored structures as corresponding to microbial cells. We will also be employing Fluorescence In Situ Hydridization (FISH) probes to determine whether these microbial cell-like structure are archaea and/or bacteria.
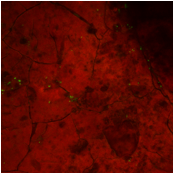
Figure 3 (image is below this legend): Confocal image of a chip surface from the same region of a GRCO shale rock stained via a LIVE/DEAD® BacLight™ bacterial viability kit. SYTO®9 green fluorescent nucleic acid stained bodies indicate presence of microbial cells whose integrity remains uncompromised. Propidium Iodide dye (red) appears to adhere non-specifically to the chip surface. This serves as a counterstain to illuminate surface features of the chip.
3b] Suitability of ATP (Adenosine Triphosphate) to determine microbial biomass in shale: This has involved quantifying ATP, a key metabolite as proxy for living microorganisms. We evaluated two workflows for quantifying ATP concentrations. Having identified one that demonstrated higher sensitivity, we are currently adapting this to shale matrices in our collection.