Reports: DNI254473-DNI2: Rates and Controls on Methane Oxidation Beneath Sea Ice
Brice Loose, PhD, University of Rhode Island
Methane Cycling beneath Sea Ice
Research Abstract
In the shallow marginal seas of the Arctic, seawater methane concentrations are frequently in excess of atmospheric saturation, partly caused by reduced sea-air-exchange around sea ice cover. These same regions can host rich ice-attached and water column microbial communities, including methane-oxidizing bacteria. How much methane accumulating beneath seasonal sea ice is oxidized by bacteria and how much escapes to the atmosphere?
To determine methane oxidation rates under a variety of methane concentrations, we collected seawater from beneath sea ice in close proximity to Point Barrow, Alaska. The seawater was amended with methane and incubated at 0 °C for up two six weeks from April through May, 2016. Methane concentrations and stable isotope ratios were monitored using a Picarro trace gas analyzer, equipped with a Small Sample Isotope Module (SSIM). High oxidation rates were observed in the presence of high methane concentrations, while surprisingly at lower concentrations the methane budget was zero or slightly positive. Trends in the mass balance were accompanied by respective enrichment or decrease of 13C-CH4, indicating that biological processes are responsible for these changes. Increasing microbial cell densities hint to microbial uptake of the methane as substrate and possibly a shift towards methanotrophic species in the community. While the low concentration experiments elucidate methane cycling at in situ concentrations, the high amended experiments show the potential of the communities to mitigate events of high methane release from the seabed or while the methane is caught under the sea ice cover. With a decreased sea ice cover or earlier ice breakup it is possible that more methane will be vented to the atmosphere due to decreased residence time in the water column and thus accessibility to the methanotrophic community.
Activities and fieldwork during FY 2016
The bulk of the PRF funding has been targeted toward supporting a postdoctoral research fellow at the U. of Rhode Island (URI) – Christiane Uhlig. Christiane joined URI in January, 2016. The first part of the year was spent preparing for the springtime fieldwork in Barrow, AK. Christiane familiarized herself with the molecular biology laboratory facilities at GSO. Field sampling equipment was purchased and tested.
When fieldwork began on April 3, the temperatures in Barrow were -25 °C and the wind speed was blowing steadily at 13 m/s. However, the sky was clear and visibility was good, with the exception of the persistent blowing snow. These cold and windy conditions would characterize the first half of the fieldtrip, and they resulted in a slow start to sampling. With a little refinement our preparations back at URI allowed us to adjust to the in-situ conditions and carry out a total of 5 field sampling days during the 21 day period. The intervening time was used to process samples for DNA extraction, DNA staining and photomicrography, and the analysis of both in-situ gas samples and microbial oxidation bags using a methane and carbon dioxide isotope analyzer from Picarro Incorporated.
The results of these experiments are currently being analyzed in preparation for the submission of two manuscripts for peer review. The first will be a methodological description of the potential application of carbon stable isotopes and the Picarro analyzer for methane studies. The second will be a detailed analysis of the microbial ecosystem in the Arctic seawater beneath sea ice and in the microbial oxidation experiments to determine community shifts and correlate with measured oxidation rates.
These experiments and results are the basis for a planned proposal to be submitted to the National Science Foundation for an upcoming drift experiment, known as the MOSAiC trans-Arctic yearlong drift experiment. The results have also been utilized to leverage additional funds to support C. Uhlig through September 1, 2017.
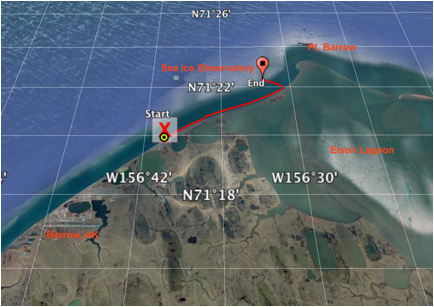
Figure 1. Google Earth Image of the field location for sea ice sampling near Pt. Barrow, Alaska.
Sampling site
The seawater used for this study was collected at two sites close to Barrow, North Slope, Alaska on 7 and 9 April 2016 in the Beaufort Sea. Site 'Elson Lagoon' (EL) was located North of Barrow (7.4.2016, 71.334 N, -156.363 W), covered with 1.5 m thick sea ice and shallow with approximately 1.5 m depth (Figure 1). Water was collected from a depth of 1.5 m in a narrow layer of water between the sea ice and the sediment. Site "Ice Mass Balance Buoy" (IMB) was located 1 km offshore of Barrow, close to the ice mass balance buoy of the sea ice physics group of University of Alaska, Fairbanks (7.4.2016, 71.373 N, -156.548 W, and 9.4.2016, 71.372 N, -156.540 W). This site was characterized by 1 m thick fast ice cover and a water depth of approximately 7 m.
During our short field sampling period we observed in situ CH4 concentrations of 61.39 nmol L-1 (n=1) at site EL and 17.22 ± 2.17 nmol L-1 (n=5) at site IMB.
Oxidation potential of the shelf bacterial community
We can extrapolate the oxidation potential determined for the under ice water bacterial community sampled in Barrow to the well-studied hotspot area for methane release in the Laptev Sea to estimate how much methane release to the atmosphere is possibly averted by bacterial oxidation under a closed sea ice cover. With oxidation rates calculated from the 50x treatments in this study during a six month ice covered period 2.500 tons CH4 would be oxidized in a water depth of 5 m in that region. In a hotspot area (0.5% of the Laptev Sea) with concentrations above 3000 nmol L-1 over a depth of 1 m, an equal amount of approximately 2.000 tonnes CH4 would be oxidized over the winter.
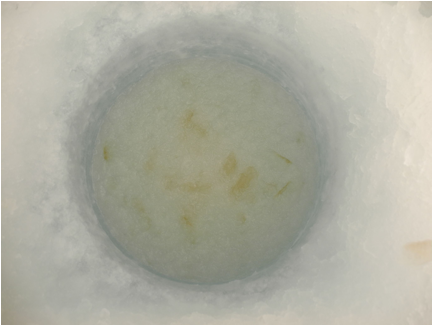
Figure 2. Ice algae observed in the under-ice slush after drilling an ice hole for under-ice sampling.
Figure 3. Sampling bags for microbial oxidation experiments being stored in a cold bath with an isothermal environmental chamber for stable oxidation conditions.