58th Annual Report on Research 2013 Under Sponsorship of the ACS Petroleum Research Fund
Reports: DNI852293-DNI8: The Use of Low-Field Nuclear Magnetic Resonance Measurements to Monitor Microbial Growth
Kristina Keating, PhD, Rutgers, the State University of New Jersey (Newark)
The focus of this research project is to determine if low-field nuclear magnetic resonance (NMR), a well-logging method used in petroleum reservoir evaluation, can be used to monitor microbial growth within geologic media. The ability to monitor microbial growth in geologic material is important for evaluating the suitability of a reservoir for Microbially Enhanced Oil Recovery (MEOR) and monitoring the progress of MEOR. This project is a three-stage laboratory study in which low-field NMR properties of microbes are characterized and the effects of microbial growth on NMR measurements monitored. Two types of NMR data sets will be used in our analysis: T2-distributions and D-T2 maps. These data sets can be used to differentiate between fluids in different pore environments (e.g. water undergoing restricted diffusion and water undergoing unrestricted diffusion); we hypothesize that these NMR measurements respond to changes in the density of microbes in the measured volume due to changes in the relative concentrations of intracellular water (i.e. water inside microbes) and extracellular water. The goal of the project is to perform fundamental research required to determine if low-field NMR measurements can monitor in situ microbial growth. This research is necessary if NMR measurements are to be used to evaluate the suitability of a reservoir for MEOR in the laboratory and to assist in the efficient application of MEOR by continuous monitoring of microbial growth in a reservoir.
In the first year of the project we focused on collecting T2-distribution measurements, using the CPMG (Carr-Purcell-Meiboom-Gill) pulse sequence, on microbial slurries and microbes in porous media both during microbial growth and on samples with known microbial density. Measurements were collected using a 2 MHz Rock Core Analyzer, which has a similar magnetic field strength to well-logging instruments. Shewanella oneidensis was used as a representative microbe in this study and clean quartz sand was used as an analog for porous media.
The NMR relaxation phenomenon results from the fact that all nuclei with an odd number of protons or neutrons possess a nuclear spin angular momentum. In a static magnetic field, the nuclear spins of a fraction of the protons will align with the static field resulting in a total magnetization that is proportional to the number of protons in the sample. In the NMR experiment a series of oscillating radio frequency (RF) pulses, tuned to perturb hydrogen spins, are applied to the sample for a short time. The excitation and return to equilibrium results in a measurable change in the bulk nuclear magnetization over time, t, described by an exponential decaying signal. For a fluid in a geological material, the echo amplitude measured using the CPMG pulse sequence, Ixy(t), exhibits multiexponential decay (Timur, 1969),
where fi is the fraction of the measured signal that relaxes with a net relaxation time of T2i. The total initial signal magnitude, Ixy0, is proportional to the total hydrogen content. The CPMG signal is typically inverted to determine a distribution of relaxation times (T2-distributions). A single relaxation time, the mean log relaxation time T2ML, is often determined from the geometric mean of the relaxation time distribution and used to represent the NMR response of geologic material.
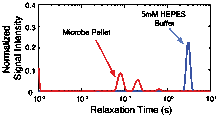
To determine the effect of microbial density on the NMR measurement microbial slurries containing a buffer solution (5mM HEPES, circumneutral pH) and S. oneidensis were created. A large vat of S. oneidensis was grown in tryptic soy broth (TSB); when peak growth had been reached, the solution was centrifuged to form a dense microbe 'pellet'. The microbe pellet was used to obtain the NMR signal from the microbes. Buffer was then added to the pellet to change the slurry density. Microbial density, in colony forming units per unit volume, was determined by calibrated optical density measurements. The T2-distribution for the microbial pellet exhibits multiple peaks and is centered at much shorter relaxation times than the T2-distribution of the buffer solution, as shown in Figure 1. Similarly, the value of T2ML decreases as a function of microbial density. These results imply that, as expected, the NMR relaxation time measurement is sensitive to the presence of microbes and the microbial density.
Figure 1: NMR relaxation time distribution of the buffer solution (5 mM HEPES, in blue) compared to the NMR relaxation time distribution of the microbe pellet (in red).
Figure 2: Mean log relaxation time versus microbial density expressed as colony forming units per volume for the NMR measurements collected on microbial slurries.
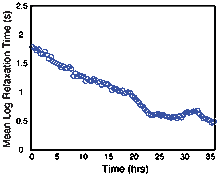
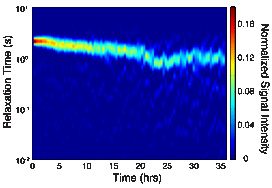
To determine the effect of microbial growth in porous media on the NMR relaxation measurement, NMR measurements were continuously collected on a column packed with pure quartz sand. TSB inoculated with S. oneidensis grown under anaerobic conditions was pumped into the columns at a rate of 0.05 mL/min. NMR measurements on the columns inoculated with S. oneidensis were compared to results from similar abiotic columns to ensure changes in the signal were due to the presence and growth of the microbes. Decreases in the NMR relaxation time and changes in T2-distribution (Figures 3 and 4) were observed in the inoculated column; the same changes were not observed the abiotic column. Changes in the NMR signal are attributed to the growth of S. oneidensis and we hypothesis that these changes are due to three factors: (1) changes in the surface area to volume ratio of the porous media due to the clogging of pores by S. oneidensis; (2) alterations in the surface properties of the porous media, causing changes in the surface relaxivity, due to the microbial activity; and (3) changes in the NMR signal due to the hydrogen present in the microbes themselves. Continued research will allow us to determine the process by which microbes affect the NMR signal.
Figure 3: Mean log relaxation time versus time for the microbes grown in synthetic porous media.
Figure 4: Relaxation time distributions versus time for microbes grown in porous media.
Copyright © 2014 American Chemical Society