Reports: ND756046-ND7: Simultaneous 3D Director Imaging and Biaxial Rheology of Lyotropic Chromonic Liquid Crystals
Itai Cohen, Cornell University
S. Zhou, S.V. Shiyanovskii, H.S. Park, O.D. Lavrentovich, Fine structure of the topological defect cores studied for disclinations in lyotropic chromonic liquid crystal, Nature Communications 8, 14974 (2017) DOI: 10.1038/ncomms14974 , 7 pages
Chenhui Peng, Yubing Guo, Taras Turiv, Miao Jiang, Qihuo Wei, Oleg D. Lavrentovich, Patterning of Lyotropic Chromonic Liquid Crystals by Photoalignment with Photonic Metamasks, Advanced Materials 29, 1606112 (2017), 6 pages
S. Zhou, O. Tovkach, D. Golovaty, A. Sokolov, I.S. Aranson, O.D. Lavrentovich, Dynamic states of swimming bacteria in a nematic liquid crystal cell with homeotropic alignment, New Journal of Physics 19, 055006 (2017), 20 pages.
Cohen published:
Liarte, Danilo B., Matthew Bierbaum, Muxin Zhang, Brian D. Leahy, Itai Cohen, and James
P. Sethna. "Visualization, coarsening, and flow dynamics of focal conic domains in simulated smectic-A liquid crystals." Physical Review E 92, no. 6 (2015): 062511.
Bierbaum Matthew, Brian D. Leahy, Alexander A. Alemi, Itai Cohen, and James P. Sethna, “Light Microscopy at Maximal Precision.” Physical Review X 7, (2017): 041007
Lavrentovich’s graduate student presented at APS March meeting:
Turiv, C. Peng, Y. Guo, Q. Wei, O. Lavrentovich, Predesigned surface patterns and
topological defects control the active matter, Abstract V16.00005, APS March Meeting, March 13-17, 2017, New Orleans, Louisiana. NSF DMR-1507637, DMS-1434185, Petroleum Research fund PRF 56046-ND7
Lavrentovich presented at:
O.D. Lavrentovich, Dynamics of living liquid crystals, Variational Models of Soft Matter, 9-13 January 2017, Pontificia Universidad Católica de Chile, Santiago, Chile.
O.D. Lavrentovich, Command of swimming bacteria by liquid crystals with topological defects and patterns, Plenary lecture, 10th Liquid Matter Conference, Ljubljana, July 17-21, 2017.
O.D. Lavrentovich, Bacterial Dynamics in Liquid Crystals, Invited presentation, XXVI International Materials Research Congress, Cancun, Mexico, August 21-24, 2017.
Cohen’s graduate student made a presentation at APS March meeting:
Leahy, Brian, Matthew Bierbaum, James Sethna, and Itai Cohen. "Measuring nm-scale Interaction Potentials with a Microscope: Light Microscopy at Maximal Precision." Bulletin of the American Physical Society 62 (2017).
Cohen presented at:
The Max Planck institute for Dynamics and Self-Organization in Gottingen, Germany.
Cornell University Physics department colloquium, Ithaca, NY, USA
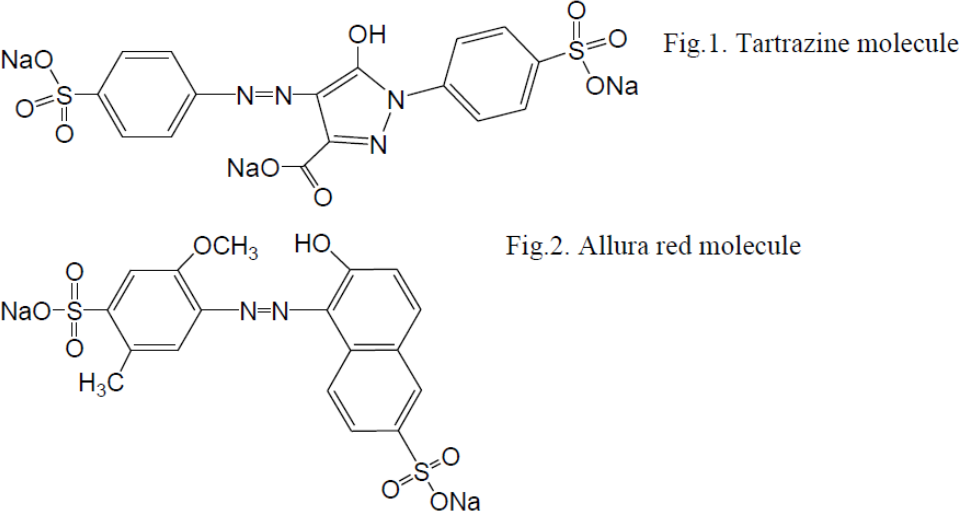
Lavrentovich determined the phase diagram of Tartrazine and Allura red. For concentration 44 wt % in water, tartrazine shows a columnar phase below 26 C; coexisting columnar and nematic phase at the temperatures between 40 C and 26 C, a coexisting range of nematic and isotropic at temperatures between 52 C and 40 C.
In contrast, Allura red in water solutions with 34 weight % concentration showed only the nematic phase, at the temperatures below 70 C; a coexisting region of the nematic and isotropic between 71 C and 88 C, and the isotropic phase above 88 C.
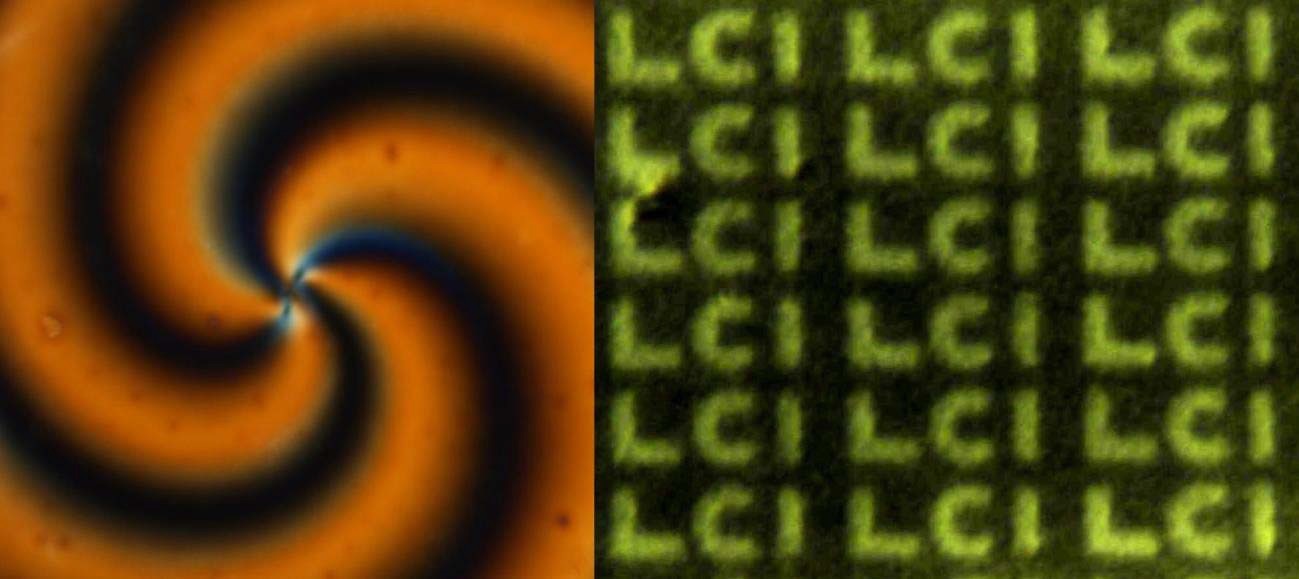
Lavrentovich developed a technique of patterned alignment of lyotropic chromonic liquid crystals. The approach is demonstrated to produce complex patterns of spatially-varying orientation of LCLCs; it requires no surfactants, and thus makes it possible to interface the patterned LCLCs with bio-matter. The demonstrated single-exposure and surfactant-free photo- patterning of LCLCs opens new opportunities in the development of advanced materials and systems based on aqueous soft matter. The patterned images made with LCLCs are shown in Fig.3.
Figure 3. Polarizing optical microscopy textures of patterned alignment of LCLCs.
These approaches were used to explore swimming of bacteria in the anisotropic environment. The director was aligned perpendicularly to the bounding plates of the cell, which is by itself an accomplishment, since the LCLCs are usually aligned parallel to the bounding plates. The paper was published in New J. Physics (2017).
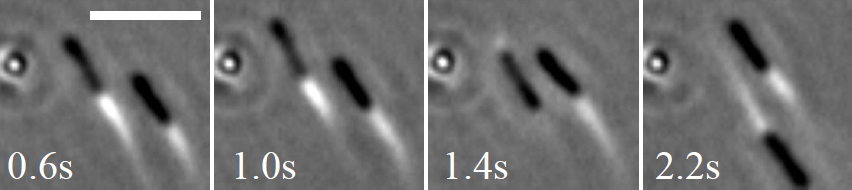
0.6s 1.0s 1.4s 2.2s
Figure 4. Swimming and backtracking of B. Subtilis perpendicularly to the director in a liquid crystal. The third bacterium is in vertical spinning state (a circle on the left hand side). Scale bar: 10 micrometers.
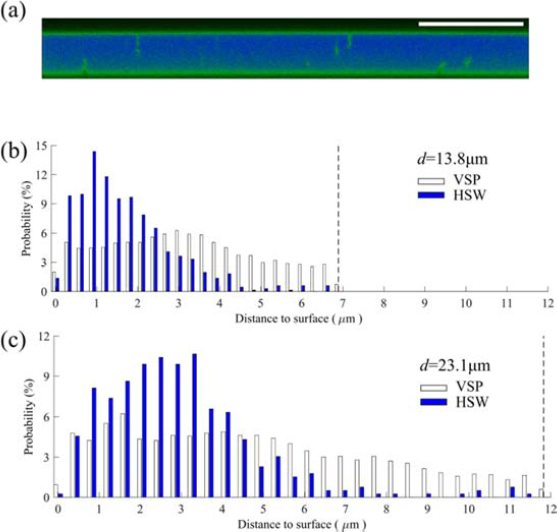
Lavrentovich also demonstrated applicability of fluorescence confocal polarizing microscopy to the studies of LCLCs. Figure 5 below shows how FCPM images allows quantification of bacteria depth distribution in LCLC cells.
Fig 5. Distribution of spinning and swimming bacteria along the z -axis normal to the glass plates.
(a) FCPM cross section of the homeotropic d = (13.8 ± 0.3) μm cell shows spinning bacteria (green vertical rods) are located close to the glass plates. Scale bar 20 μm . (b, c) Probability distribution of detecting any part of VSP and HSW bacteria versus distance to the glass plates in (b) the d = (13.8 ± 0.3) μm cell and in (c) the d = (23.1 ± 0.5) μm cell. The dashed lines mark the middle plane of the LCLC cells.
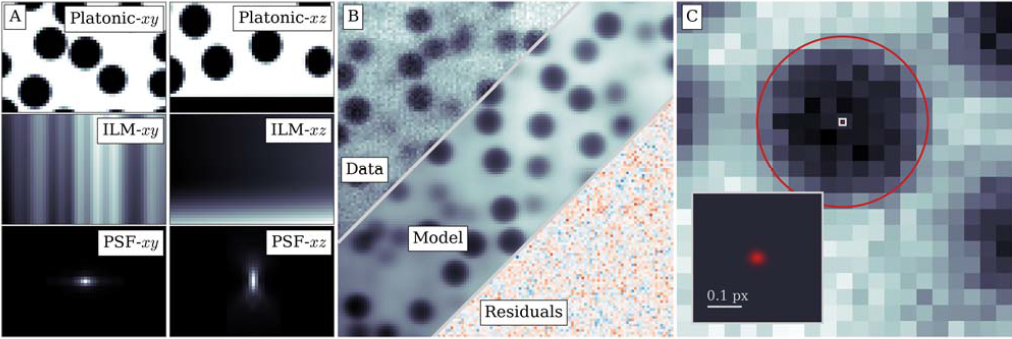
Cohen, using a generic, methodological approach, extracted the information in a confocal microscope image by fitting experimental images with a detailed optical model of the microscope, a method called parameter extraction from reconstructing images. As a proof of principle, they demonstrated this approach with a confocal image of colloidal spheres, improving measurements of particle positions and radii by 10–100 times over current methods and attaining the maximum possible accuracy. With this unprecedented accuracy, they measured nanometer-scale colloidal interactions in dense suspensions solely with light microscopy. The approach is generic and applicable to imaging methods from brightfield to electron microscopy, where accuracies of 1 nm and 0.1 pm, respectively are expected.
FIG. 1. Information recovered from real confocal microscope images. (a) The generative model consists of a platonic image of dye distributed around perfect spheres and a coverslip (top panels), illuminated with a spatially varying intensity (middle panels), and convolved with a physical point-spread function (bottom panels). The left panels show the model components, and the right panels show the combined image. (b) These components combine to form a realistic generative model (bottom right), which, aside from noise, is visually indistinguishable from the data (top left). (c) From the fit parameters, we extract information such as particle positions and radii (orange highlights) to within a few percent of a pixel–corresponding to an accuracy of 1 and 3–4 nm, respectively. The inset zooms into the center of the particle (white square), highlighting the precise localization and calculated uncertainty of the particle’s position.