Reports: ND255615-ND2: Roles of Microbial Hydrolytic Enzymes in Mediating Carbon Cycling in Shale Formations of the Southwestern United States
Farooq Azam, PhD, University of California, San Diego
I] Impact of this grant on:
1] Research directions in the laboratory of the Principal Investigator:
My research interests in microbial oceanography have had an increasing emphasis on microbial mediated carbon cycling. My work primarily focuses on the marine water column. I am interested in studying oceanic micro-scale patchiness to be able to extrapolate such insights to the ecosystem level and thereby arrive at a better understanding of the role of microorganisms in transforming different carbon pools in the marine water column. Studying such processes is critical to predict the fate of carbon dioxide in the world's oceans. On the other hand, massive sinks of organic matter and microbial load in sediments below the ocean floor are not as well constrained (this has implications for release of C from such deposits). This is the case even for large shale formations in the United States. This grant has enabled me to pursue a new line of investigation involving shale geo-microbiology with new collaborators while also drawing on tools and concepts developed in my laboratory.
2] Training and career development
This grant has provided a platform for training personnel at different career stages. Dr. Subbarao has a keen interest in geo-microbiology and he has continued to work on this project in its second year. Dr. Subbarao has made excellent progress and plans to synthesize with me the ongoing work into a manuscript in this coming year. This grant has enabled Dr. Subbarao to:
i] Gain experience employing a range of concepts and protocols spanning the molecular to cellular scales in geo-microbiology.
ii] Initiate and maintain collaborations nationally and internationally. Our geoscience collaborators have enabled us to undertake a multidisciplinary characterization of shale samples that would otherwise have been impractical.
iii] Participate in fieldwork to collect diverse shale samples.
iv] Train undergraduate and masters level students in our project. These students would hopefully be enthused by exposure to such a multidisciplinary project, motivating them to consider careers in scientific research.
II] Work progress:
1] Sample collection and processing: Dr. Subbarao participated in a geological field trip to northern Colorado with Prof Manika Prasad from the Colorado School of Mines (CSM) in late 2016. Prof Prasad who is an expert in shale geology and geophysics guided Dr. Subbarao in the field. Samples were collected in a manner compatible for downstream microbiological analysis. Prof Prasad also provided other select shale specimens for our analysis from her sample repository.
2] Characterizing Microbial diversity of samples from shale formations in the western United States: We have adopted two complementary culture-independent approaches (below) to study the diversity of microbial communities in our selected shale sample set.
2a] 16S rRNA gene sequencing: Genomic DNA was extracted from a range of shale samples, some of which were rather challenging. We are currently analyzing Illumina tag sequence data to determine microbial diversity from our shale samples as well as infer trends of microbial diversity across these samples spanning provinces, geological age and organic matter content.
2b] Intact Polar Lipids (IPLs): These membrane lipid molecules are used in a chemotaxonomic approach to determine microbial diversity in sedimentary matrices of varied shale samples. The advantage of IPL analysis over gDNA based 16S rRNA gene sequencing is the ability to detect microorganisms that are 'viable' as distinct from the those that are fossilized, as well as determining the ratio of archaea to bacteria in the samples. Our collaboration on this theme with Dr. Julius Lipp at the University of Bremen is ongoing with Dr. Subbarao making a productive visit to Bremen this summer to work on assignment of mass spectrometry data. Archaeal IPL data has been processed to yield a distribution of lipid classes (which in turn are derived from specific archaeal classes) while bacterial IPL data is currently being analyzed.
3] Presence and nature of microorganisms in shale matrices:
We have built on progress of this theme that was based on confocal laser scanning fluorescence microscopy in year 1 of this project. This work is ongoing with various shale rocks. We discovered the presence of either 'live' individual microbial cell-like bodies or their congregations along fractures/crevices on internal surfaces of shale rocks. This prompted us to investigate the morphology, size/shape distribution and topology of presumed microbial community clusters at higher magnification and resolution by scanning electron microscopy (SEM).
3a] High resolution imaging of shale surfaces via SEM: Dr. Subbarao set up a collaboration with Dr. Mark van Zuilen at the University of Paris to image shale samples (surfaces) with SEM. Preliminary analysis of SEM images suggests that microbial cell like bodies that could be discerned by confocal microscopy do possess a range of shape (rods/ spheres) and size characteristics typical of microorganisms. Even cell-like bodies that seem to be dividing can be visualized in some images as also cellular bodies with lengths as low as 0.4 mm (Figure 1 from a California shale sample at 15.53X magnification). We plan to perform elemental composition of these bodies by EDX for further evidence of their biogenic nature.
Fig 1: Microbial like cluster in the center of this SEM image
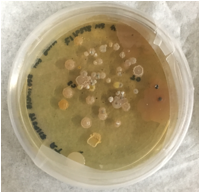
3b] Culturing microorganisms from shale samples: We have been successful in culturing microorganisms from select shale samples (Figure 2). We are currently determining the phylogenetic identity of these isolates by 16S rRNA gene sequencing. We intend utilizing these isolates under controlled conditions to test for their ability to hydrolyze and utilize shale associated organic matter.
Fig 2: Microbial colonies from a Colorado shale sample
3c] Tracking metabolites in select microbial isolates from shale matrices:
We initiated a collaboration with Dr. Abdesh Kumar at Scripps Center for Marine Biotechnology and Biomedicine to determine the metabolites produced by shale derived microbial isolates. Initial liquid chromatography-Mass Spectrometry data indicate presence of microbial metabolites. Detailed metabolome analysis is underway and we hope that this will provide insights on microbe-organic matter interactions within the shale matrices.