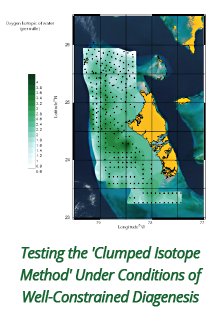
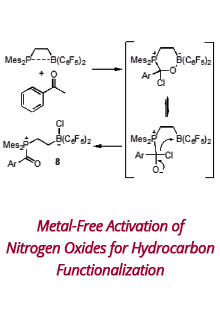
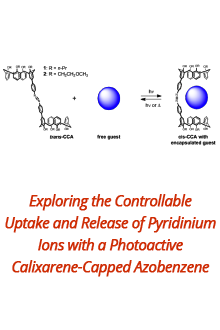
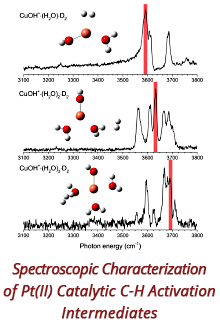
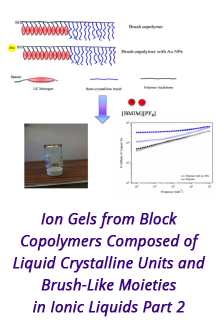
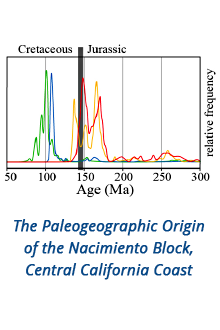
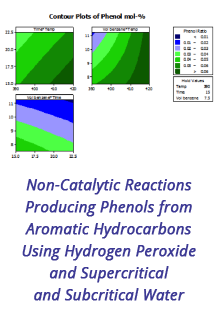
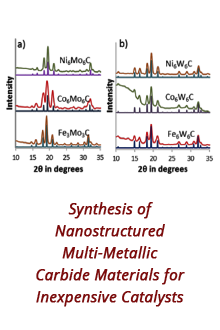
Reports: DNI1052404-DNI10: Validating Computational Design Principles for Crystalline Enzyme Assemblies
Christopher D. Snow, Ph.D., Colorado State University
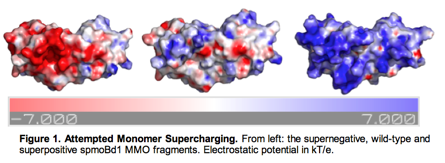
Monomer Redesign for Solubility
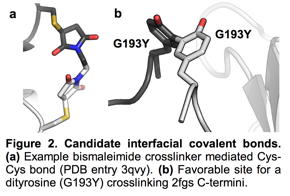
Optimizing Adventitious Crosslinking
Efficient Crystal Search Algorithm in Hexagonal Space
Groups