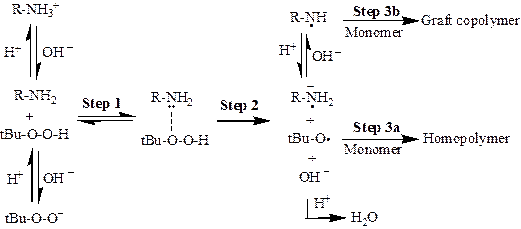57th Annual Report on Research 2012 Under Sponsorship of the ACS Petroleum Research Fund
Reports: DNI750942-DNI7: Solvent-Assisted Coalescent Assembly of Amphiphilic Core-Shell Nanoparticles
Junmin Zhu, PhD, Case Western Reserve University
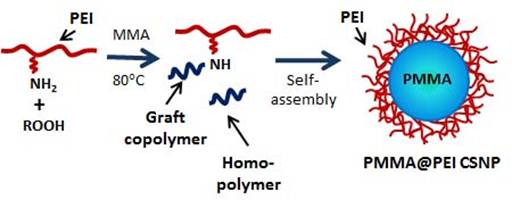
The assembly of nanoparticles is a key step for the development of advanced nanodevices with various structural designs for fundamental interface studies, industrial and biomedical applications. However, the major impediments to progress in the development of advanced nanomaterials are the lack of versatile nanoparticles as building blocks, undefined control of assembly processes, and limited potential for scale-up production. To address these challenges, the proposed research seeks to develop a novel strategy, solvent-assisted coalescent assembly of core-shell nanoparticles (CSNPs) into advanced nanostructures. Hydroperoxide-amine initiated graft polymerization (HAIGP) provides a facile way to prepare well-defined amphiphilic CSNPs with hydrophilic amine-containing polymers (HAPs) like branched polyethylenimine (PEI) as the shell and hydrophobic poly(vinyl monomer) like poly(methyl methacrylate) as the core (Fig 1). These unique CSNPs have hydrophobic homopolymers in the core, which can be extracted to form hollow nanoparticles or assembled by coalescence into nanaofibers or nanotubes. Over the first year of the project, our work has focused on two aspects, including (i) pH effects on hydroperoxide-amine initiated graft polymerization (HAIGP) for synthesizing PMMA@PEI CSNPs, and (ii) assembly of PMMA@PEI CSNPs into advanced nanostructures by a solvent-assisted coalescence method.
Fig 1. Process of HAIGP to fabricate PMMA@PEI CSNPs
pH effects on HAIGP to synthesize CSNPs. The pH effects on HAIGP is shown in Fig 2. The amine groups on HAPs such as polyethylenimine (PEI) can act as electron donors to form redox initiation pairs with alkyl hydroperoxide (ROOH) like tert-butyl hydroperoxide (TBHP), which results in the formation of HAP macroradicals and alkoxyl radicals. HAP macroradicals initiate the graft of vinyl monomers like methyl methacrylate (MMA) from HAPs, while the alkoxyl radicals initiate the homopolymerization of vinyl monomers. Graft copolymers and homopolymers are generated concurrently. HAPs with growing hydrophobic chains generated in situ, not only self-assemble with the homopolymers to produce micelle-like microdomaines, but also act as both electrostatic and steric stabilizers to provide stability to the developing nanoparticles. Therefore, the process can fabricate CSNPs with high solid contents (up to 20%, w/w), without adding surfactants. Since PEI contains many amine groups, which are easy to be protonated, pH plays an important role in affecting the redox pair formation between amine groups and TBHP, graft copolymer formation and particle size of CSNPs.
Fig 2. pH effects on HAIGP
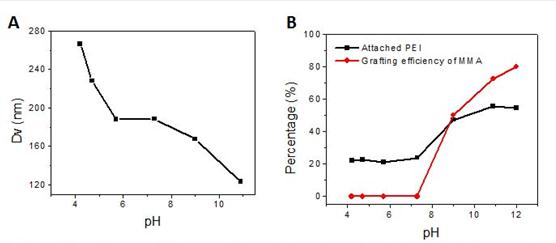
To investigate the pH effects on HAIGP, PMMA@PEI CSNPs were synthesized by using branched PEI(Mw 60K) and MMA (1:4, wt%/wt%) with 0.1mM TBHP and reacting at 80°C for 2h under Ar and magnetic stirring at 600 rpm. The pH of the aqueous phase was adjusted from 4.0 to 11.0 by adding 1M HCl before adding MMA and TBHP. The results show that when reducing the pH (more acidic), the particle size increased (Fig 3A), while the MMA conversion and PEG attachment decreased, due to the protonation of the amine groups on PEI, resulting a more PEI swelling structure and less redox pair formation. Generally, the particle size ranged between 100 to 300 nm with 80% to 90% MMA conversion and 20-55% of PEI attachment. The MMA grafting efficiency is about 80% at pH 11, while it dropped very low (~10%) at pH < 7.0 (Fig. 3B).
Fig 3. pH effect on particle size (A) and MMA conversion and PEI attaching percentagy (B) of PMMA@PEI CSNPs.
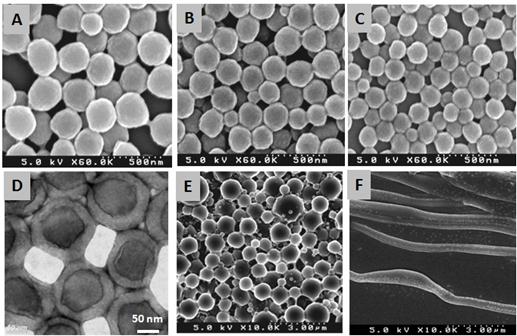
The pH effect on particle size was also observed by SEM, as shown in Figs 4A-C. The average particle sizes are 250nm, 200nm and 150nm at pH=4.0, 7.0 and 11.0, respectively. Thus, when decreasing the pH, the particle size becomes greater due to the protonation of amine groups on PEI. Also, the core-shell structure of the nanoparticles formed by HAIGP at pH=11 was confirmed by TEM analysis (Fig 4D). The outer layer is hydrophilic, consisting of PEI, while the core is hydrophobic, consisting of PMMA.
Fig 4. SEM images of PMMA@PEI CSNPs prepared at pH= 4.0 (A), 7.0 (B), and 11.0 (C). TEM image of PMMA@PEI at pH =11.0 (D), and chloroform-assisted assembly of PMMA@PEI CSNPs prepared at pH=7.0 (E) and pH =11.0 (F).
Solvent-assisted assembly of CSNPs. PMMA@PEI CSNPs have an unique structure feature with homopolymer PMMA in the core, which can be dissolved and extracted by organic solvents to form hollow structures. Our results show that the addition of organic solvents into latex PMMA@PEI CSNPs was able to overcome the core/shell interfacial energy barrier to induce the interparticle interactions under shear flow, while controlling of the solvent solubility and water miscibility assisted the coalescent assembly of CSNPs into advanced nanostructures, such as nanofibers and nanotubes. Chloroform was used for solvent-assisted coalescent assembly of CSNPs by an extraction and evaporation method. The PMMA@PEI CSNPs prepared at pH 7 (with low graft copolymers) assembled into bigger spherical particles with a diameter of about 1mm (Fig 4E), while the same CSNP samples prepared at pH 11 (with higher graft copolymers) assembled into nanofibers with a diameter of 300 nm (Fig 4F). This may be due to the difference in the concentration of graft copolymers. More research on the controlling of CNSP assembly is in progress.
Impact of the research. The support of this ACS PRF grant has enabled the PI to develop HAIGP as a novel strategy to prepare CSNPs, establish a versatile coalescence method for assembling CSNPs into advanced nanostructures, and collect key results for preparing two manuscripts and one NSF proposal (to be submitted this year). In addition, one postdoc and two undergraduate researchers have been supported by this award, and they have learned valuable skills in the synthesis and characterization of polymers and nanomaterials, graft emulsion polymerization, and molecular self-assembly.