

44410-G3
Understanding the Origin of Suicide Inactivation in the Extradiol Dioxygenases
As described in the previous progress report, we have focused our efforts on the studies of the hydroquinone (HQ) ring-cleaving dioxygenases and Fe(II)-containing model complexes designed to mimic these enzymes. The preliminary results described below (and in the previous progress report) were used in an NSF grant that was submitted in July, 2008.
1.
Several Fe(II)-containing ring-cleaving dioxygenases are homologous to extradiol dioxygenases, but operate on very different substrates and are poorly characterized. One is 2,6-dichlorohydroquinone 1,2-dioxygenase (denoted here by the gene name, PcpA) from Sphingomonas chlorophenolica. Unlike the extradiol dioxygenases, where few enzymes can cleave chlorinated substrates successfully, for PcpA, 2,6-dichloro-HQ is the native substrate, and it exhibits no evidence of suicide inactivation.
In the previous report, we described using an express system for PcpA obtained from Prof. Luying Xun. We have scaled up our expression of PcpA in order to pursue crystallization trials and other more detailed characterization. The current vector has both a His-tag and an S-tag at the N-terminus. The His-tag has a thrombin cleavage site, and thrombin digestion easily removed this tag. We performed crystallization screens have obtained some promising leads. However, better success will likely be obtained by removal of both tags. The S-tag has an enterokinase cleavage site; however, we found that enterokinase could not efficiently cleave the tag without causing degradation of the protein. We performed site-directed mutagenesis to change the enterokinase cleavage site to a thrombin cleavage site, and are screening growth conditions of this new vector for optimal expression.
In the previous report, we
described the homology-based structural model of PcpA that we generated based
upon a crystal structure of a putative bacterial glyoxalase that contains a Zn(II)
center (denoted here by its PDB number, 1ZSW).
We obtained an expression system for 1ZSW from Prof. Wayne Anderson in
order to test its enzyme activity. We expressed
the protein in high yield, purified it by metal-affinity chromatography, and
tested its enzymatic activity. 1ZSW
exhibited no glyoxalase activity in the presence of Zn(II), Ni(II), or Co(II). We screened 1ZSW for oxidative ring-cleavage
ability with Fe(II) and with the substrates listed above. It did exhibit extremely weak ring-cleavage activity
with 3-methylcatechol. Although we were unable to produce enough ring-cleavage
product to examine by techniques other than UV/visible absorption spectroscopy,
this spectrum was consistent with predominantly 1,6-cleavage of the substrate. This indicates that 1ZSW is not a glyoxalase,
but is mostly likely a ring-cleaving dioxygenase, although the native substrate
is not known. Note that there are
>200 close homologues of 1ZSW in the NCBI database, all of unknown function.
Another known HQ dioxygenase is from
Sphingomonas paucimobilis UT26 (gene
name LinE). Unlike PcpA, LinE has been reported
to be active towards HQ and 2-chloro-HQ. We obtained the bacterium Sphingobium indicum CCM 7286, which has a LinE gene identical to
that of S. paucimobilis UT26. We grew
this organism, extracted its genomic DNA, and successfully cloned the LinE gene
into a pET-28a vector, which has an N-terminal His-tag with a thrombin cleavage
site. We are currently screening conditions
for optimal expression.
2. In collaboration with Prof.
Patrick Holland at the
 We
have been performing more detailed steady-state kinetic measurements of PcpA in
order to determine its substrate specificity. We screened HQ, 2-chloro-, 2-bromo-,
2-methyl-, 2,5-dichloro-, 2,6-dibromo- and 2,6-dimethyl-HQ; catechol, 3-methyl-
and 4-methylcatechol; 2,6-dichloro-, 2,6-dibromo-, and
4-amino-2,6-dichlorophenol; and gentisate and pyrogallol. Only 2,6-dibromo- and 2,6-dimethyl-HQ showed
evidence of ring cleavage. Surprisingly,
2,6-dibromo-HQ exhibits essentially the same kinetic parameters as the native
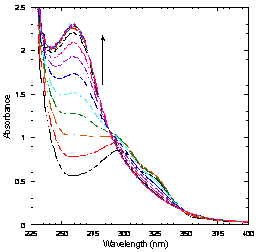
substrate. A UV/visible spectrum showing
ring cleavage of this substrate is shown to the right. 2,6-dimethyl-HQ is a much poorer substrate,
and we are working to characterize the products of the reaction with this
substrate. We have also tested to see if
any of these molecules act as inhibitors.
Preliminary evidence suggests that 2-bromo-HQ is a strong inhibitor and
2-chloro- and 2-methyl-HQ are weaker inhibitors.
We
have been performing more detailed steady-state kinetic measurements of PcpA in
order to determine its substrate specificity. We screened HQ, 2-chloro-, 2-bromo-,
2-methyl-, 2,5-dichloro-, 2,6-dibromo- and 2,6-dimethyl-HQ; catechol, 3-methyl-
and 4-methylcatechol; 2,6-dichloro-, 2,6-dibromo-, and
4-amino-2,6-dichlorophenol; and gentisate and pyrogallol. Only 2,6-dibromo- and 2,6-dimethyl-HQ showed
evidence of ring cleavage. Surprisingly,
2,6-dibromo-HQ exhibits essentially the same kinetic parameters as the native
substrate. A UV/visible spectrum showing
ring cleavage of this substrate is shown to the right. 2,6-dimethyl-HQ is a much poorer substrate,
and we are working to characterize the products of the reaction with this
substrate. We have also tested to see if
any of these molecules act as inhibitors.
Preliminary evidence suggests that 2-bromo-HQ is a strong inhibitor and
2-chloro- and 2-methyl-HQ are weaker inhibitors.
 Fe(II)-containing model complexes with
non-catechol substrates
Fe(II)-containing model complexes with
non-catechol substrates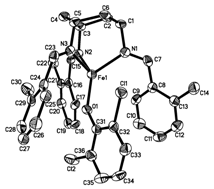 Addition
of the following deprotonated substrates to a 1:1 (TACH-o-tol)-Fe complex in acetonitrile-d3 yielded an immediate color change and the appearance of
the paramagnetically shifted 1H NMR spectra: phenol, 2-chlorophenol,
2,6-dichlorophenol, 2-aminophenol and 2-methyl-HQ. These data indicate formation of 1:1:1
complexes containing TACH-o-tol,
Fe(II) and deprotonated substrate. Similar results were obtained with the TACH-cinn
ligand. We have obtained a crystal
structure of the complex
Addition
of the following deprotonated substrates to a 1:1 (TACH-o-tol)-Fe complex in acetonitrile-d3 yielded an immediate color change and the appearance of
the paramagnetically shifted 1H NMR spectra: phenol, 2-chlorophenol,
2,6-dichlorophenol, 2-aminophenol and 2-methyl-HQ. These data indicate formation of 1:1:1
complexes containing TACH-o-tol,
Fe(II) and deprotonated substrate. Similar results were obtained with the TACH-cinn
ligand. We have obtained a crystal
structure of the complex