44382-G4
Thiamine-Dependent Decarboxylation Reactions in Methanogenic Coenzyme M Biosynthesis
Methanogens produce more than 400 Tg of methane each year, corresponding to 80% of global methane production from all sources. This methane is a valuable energy source, as well as a greenhouse gas. To develop new ways of controlling methanogenesis, we are elucidating the biosynthetic pathways for two coenzymes that these archaea require to produce methane. Coenzyme M (CoM; 2-mercaptoethanesulfonate) is the terminal methyl carrier. Coenzyme B (CoB; containing 7-mercaptoheptanoyl-threonine phosphate) reduces the methyl-CoM thioether bond in a reaction catalyzed by the methyl-CoM reductase enzyme. Treatments that block the biosynthesis of CoM or CoB could be selective inhibitors of methanogenesis.
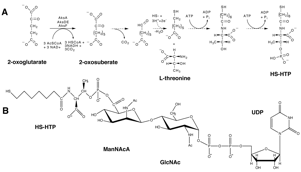
We previously identified the pathway that Methanocaldococcus jannaschii uses to make CoM (Fig. 1A). The third
enzyme in this pathway uses a thiamine pyrophosphate (TPP) coenzyme to catalyze
the decarboxylation of sulfopyruvate, producing sulfoacetaldehyde. We have
developed a system to express and purify soluble sulfopyruvate decarboxylase
enzyme (ComDE) from the methanogen Methanosarcina
acetivorans. We have shown that high TPP levels are required for ComDE
activity in the absence of dithionite. Ongoing studies will determine whether
TPP acts as a cofactor or substrate in these reactions, and why this enzyme is
inactivated by oxygen. The genome sequence of Methanosarcina
acetivorans has no homologs of the comA,
comB or comC genes that we identified in M. jannaschii. In this project, we have recently identified a new
pathway for CoM biosynthesis in this microorganism that uses phosphoserine to
make cysteate, which can be transaminated to produce sulfopyruvate (Fig. 1B).
From this methanogen, we have cloned and expressed a novel enzyme, cysteate
synthase, that evolved from the threonine synthase enzyme. This enzyme
catalyzes the elimination of phosphate from phosphoserine and the nucleophilic
addition of sulfite to a dehydroalanine intermediate, forming L-cysteate. Mass
spectral data and phosphate analyses confirm this activity. We previously
described the methanogen aspartate aminotransferase that efficiently produces
sulfopyruvate from cysteate. These experiments
demonstrated that sulfopyruvate biosynthesis evolved convergently in the Methanococcales and in the Methanosarcinales lineages.
Consequently, the sulfopyruvate decarboxylase protein is the best target for
designing a broad-spectrum inhibitor of methanogen CoM biosynthesis. Figure 2. Proposed pathway for the biosynthesis
of HS-HTP and an extended form of CoB. In contrast to CoM, CoB has only been found in methanogens. The structure of CoB was
originally reported to be 7-mercaptoheptanoylthreonine phosphate (HS-HTP) (Fig.
2A). A subsequent report identified a UDP-disaccharide headgroup bound to
HS-HTP through a labile phosphoanhydride bond (Fig. 2B). In this project, we
identified and characterized all of the enzymes from Methanococcus maripaludis that are required to make the nucleotide-sugar
precursors for this headgroup: UDP-N-acetylglucosamine
(GlcNAc) and UDP-N-acetylmannosaminuronate
(ManNAcA). These enzymes will be valuable tools to make substrates for future
studies of CoB biosynthesis. Other enzymes identified in this project have
already proven useful for the analytical determination of UDP-N-acetylglucosamine concentrations in
cells. The heptanoate moiety of CoB is produced by a series of
2-oxoacid elongation reactions, which extend 2-oxoglutarate to 2-oxosuberate
(Fig. 2A). The three enzymes that catalyze these reactions evolved from the
isopropylmalate pathway that most organisms use for leucine biosynthesis. We
have identified and characterized the two hydro-lyase enzymes from M. jannaschii that have isopropylmalate
isomerase (IPMI) and homoaconitase (HACN) activities. Through this project, we
identified the first homoaconitase enzyme that catalyzes both half-reactions
–completely converting (R)-homocitrate
to (2R,3S)-homoisocitrate. This iron-sulfur protein efficiently binds all
of the different chain length analogs that are intermediates in the three
cycles of 2-oxoacid elongation shown in Fig. 2A. PRF funding for this project has benefitted a number of
graduate and undergraduate students, as well as one ACS Project SEED high
school student. By providing supplies and stipends this grant has helped these
students pursue independent research in microbial biochemistry and develop
skills in problem solving and experimental design. This grant has provided
valuable startup funds to the PI, who has garnered competitive grants from NIH
and NSF based on preliminary data from this work.
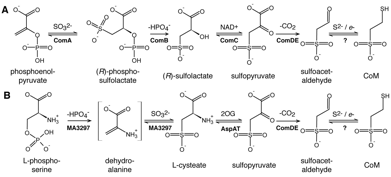 Figure 1. Biosynthesis of CoM in the Methanococcales (A) and the Methanosarcinales (B).
Figure 1. Biosynthesis of CoM in the Methanococcales (A) and the Methanosarcinales (B).