
ACS PRF | ACS
All e-Annual Reports

44410-G3
Understanding the Origin of Suicide Inactivation in the Extradiol Dioxygenases
1. We obtained 4,4'-difluoro-2,3-dihydroxybiphenyl (DFDHB) and Sphingomonas RW1 2,2',3-trihydroxy-biphenyl dioxygenase (THBD) from our collaborator, Prof. Lindsay Eltis (U. British Columbia). The pH titration of aqueous DFDHB monitored by 19F-NMR (Fig. 1, A and B) revealed a pKa of ~8.2. When DFDHB was bound to the enzyme, two 19F peaks were observed, one peak was sharp and shifted slightly relative to aqueous DFDHB (distal fluorine), the other shifted and broadened dramatically (proximal fluorine). The pH titration of DFDHB bound to the enzyme monitored by 19F-NMR did not reveal any significant shifts in the positions of either peak over the pH range of 8.2-9.7 (Fig. 1D), indicating that the substrate remains bound as the monoanion over this pH range. Preliminary kinetic data indicates that DFDHB is a potent suicide inactivator of THBD. We plan to prepare other substituted 2,3-dihydroxybiphenyls and compare results among these different substrates. 2. Synthesis of the ligand described in the Proposal proved difficult and was not pursued. Bruijnincx et al reported ligands that provide the desired N, N, O binding framework, the 3,3-bis(1-alkylimidazol-2-yl)propionate ligand family. In collaboration with Prof. Patrick Addition of 3,5-di-tert-butylcatechol and triethylamine to solutions containing LFe(II)(H2O)3 and L2Fe(II) yielded lavender-colored solutions with no 1H NMR signals other than those from triethylamine and solvent. A purple solid was be obtained that has not been structurally characterized. Bruijnincx et al reported similar results, again without structural characterization. Our studies indicate that this species is probably not a monomeric Fe(II) complex. We have begun pursuing other ligand designs that will better allow us to obtain the desired Fe(II)-catechol model compounds. 3. Studies of 2,5-dichloro-p-hydroquinone dioxygenase Several Fe(II)-containing ring-cleaving dioxygenases are homologous to extradiol dioxygenases, but operate on very different substrates and are poorly characterized. One is 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (denoted here by the gene name, PcpA) from Sphingomonas chlorophenolica. Unlike the extradiol dioxygenases, where few enzymes can cleave chlorinated substrates successfully, for PcpA, 2,6-dichloro-p-hydroquinone is the only substrate that is cleaved appreciably, nor is there evidence of suicide inactivation. Given this, and preliminary evidence that it is suitable for paramagnetic NMR, characterization of this enzyme became a high priority. Prof. Luying Xun ( Mutants of PcpA were constructed in which each of the putative ligands (H11, H159, H227, and E276) were mutated to alanine. A fifth mutant was constructed in which Y266 (a putative second-coordination sphere residue) was mutated to phenylalanine. All five mutants were expressed and purified to homogeneity. Preliminary kinetic data indicates that all of the mutants are inactive except for H159A, thus supporting the active site predicted by the homology-based structural model. Since we can express and purify PcpA in high yield, and given its the unique substrate specificity, it is an ideal system with which to study the effect of chloro- substituents on ring-cleaving dioxygenase activity. We plan to devote considerable attention to this system in the coming funding cycle. 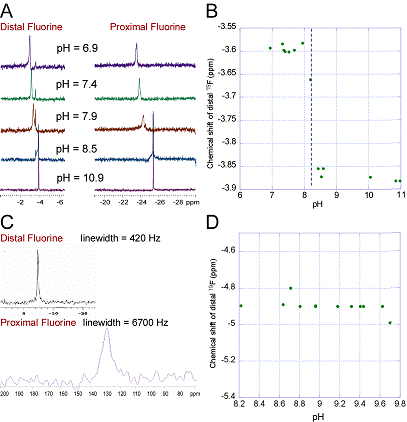 Determination of the pKa of substrates bound to an extradiol dioxygenase
Determination of the pKa of substrates bound to an extradiol dioxygenase Preparation and characterization of model complexes of extradiol dioxygenases
Preparation and characterization of model complexes of extradiol dioxygenases Previous work on PcpA indicated that the ligands to the Fe(II) center were the same as found in Type-1 extradiol dioxygenases, based upon primary sequence homology: H159, H227, and E276. Recently, a crystal structure was reported of a putative bacterial glyoxalase (denoted here by its PDB number, 1ZSW) with a higher sequence homology to PcpA than any extradiol dioxygenase. 1ZSW contains a Zn(II) center ligated by H11, H227, and E276 (PcpA numbering). From this structure, we generated a homology-based model of the structure of PcpA (Fig. 3), which predicted that the metal-binding site of PcpA should be similar to that of 1ZSW, while H159 should be distant from the active site.
Previous work on PcpA indicated that the ligands to the Fe(II) center were the same as found in Type-1 extradiol dioxygenases, based upon primary sequence homology: H159, H227, and E276. Recently, a crystal structure was reported of a putative bacterial glyoxalase (denoted here by its PDB number, 1ZSW) with a higher sequence homology to PcpA than any extradiol dioxygenase. 1ZSW contains a Zn(II) center ligated by H11, H227, and E276 (PcpA numbering). From this structure, we generated a homology-based model of the structure of PcpA (Fig. 3), which predicted that the metal-binding site of PcpA should be similar to that of 1ZSW, while H159 should be distant from the active site.