Reports: AC4
44160-AC4 Investigation of Dye-Protein Interactions and Optimization of Fluorescence-Based Assays for Target Binding of Calmodulin
Many modern experimental techniques are based on fluorescence detection. For studies of proteins, the application of these techniques requires labeling the protein with visible fluorescent dyes. The objective of this project is to understand how dyes interact with proteins and how this interaction affects fluorescence measurements and specifically fluorescence polarization (FP) methods. In FP, binding of a smaller molecule to a larger one is detected by the change in fluorescence depolarization that results from the change in volume of the rotating species. The sensitivity of this measurement depends critically on the interaction or lack of interaction of the fluorophore with the protein, i.e. does the dye molecule stick to the protein or does it move freely in solution, tethered to the protein like a balloon on a string.
As the model protein for these
studies we have chosen the calcium signaling protein calmodulin (
Surface Adsorption
For FP competition measurements
involving CaM-binding peptides, we labeled the peptide M13, a CaM-binding
peptide, with the dye Atto465. We previously
reported an FP measurement
Figure 1. Influence of the presence of surface modifying agents on the change in fluorescence intensity of M13-Atto465. Fluorescence intensities were measured in standard high Ca2+ HEPES buffer at pH 7.4 and 25 °C in a polystyrene cuvette. The fluorescence intensity at each time interval is an average of 10 second time trace collected at 510 nm by exciting at 453 nm.
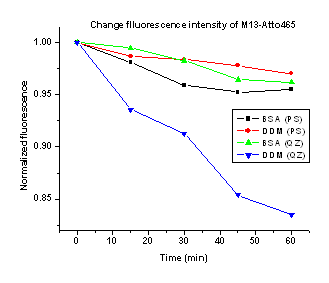
In the presence of BSA, however, the FP of M13-Atto465 is
high, suggesting interaction of M13 with BSA. In contrast, DDM does not lead to high
FP. Thus, DDM should be useful to
prevent surface adsorption. However, we
wondered whether DDM would interfere with
|
% of DDM present
|
Dissociation constant (± 0.07 nM) |
|
0
|
0.30
|
|
0.005
|
0.24
|
|
0.01
|
0.10
|
|
0.02
|
0.13
|
Fluorescence Polarization Assay
The competitive
the dissociation constants of competing ligands can
be measured without labeling each of the target ligands.
We have developed a competitive FP binding
assay to measure the affinity of ligands for
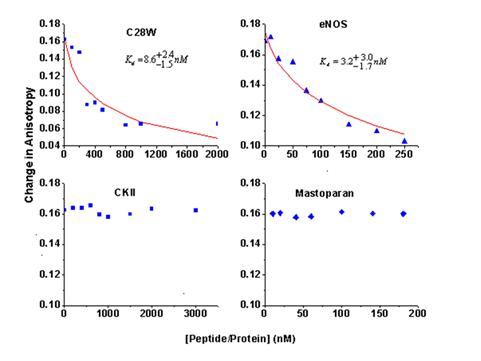
Figure 2. The change in fluorescence anisotropy of M13-Atto65 bound to CaM upon competitive ligands binding to